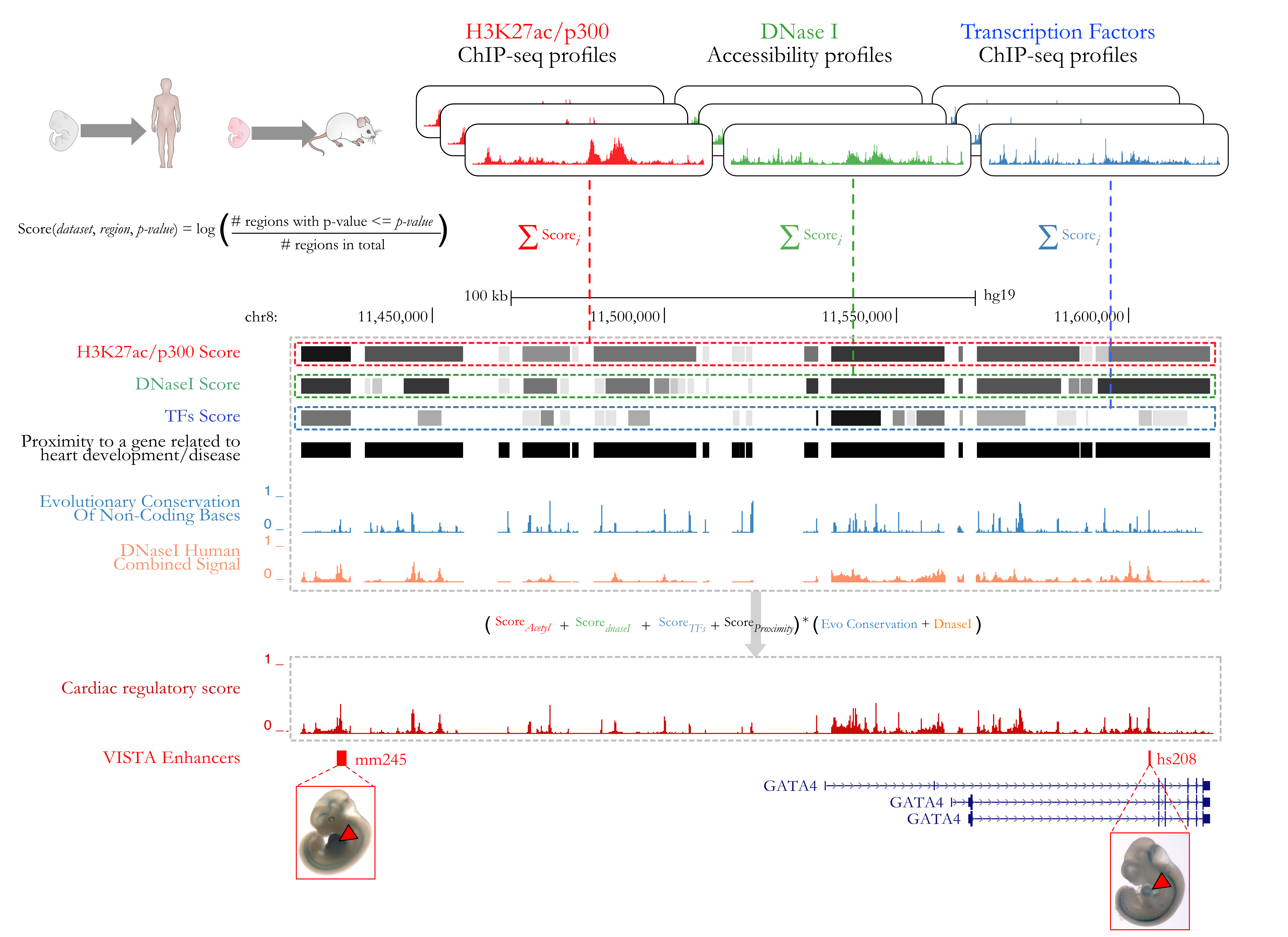
This session contains the compendium of >80,000 predicted human heart enhancers described in Dickel et al. (2016) Nature Communications. Enhancer predictions were generated by performing an integrative analysis on >35 epigenomic datasets (H3K27ac and p300/CBP) from human and mouse heart tissue including: prenatal human heart, major anatomical subregions of childhood and adult human hearts, and a closely spaced developmental time series of prenatal and postnatal mouse heart. Each predicted enhancer is scored 0 (weak) to 1 (strong) to indicate the epigenetic support for the prediction.
We have since enhanced this compendium to generate a cardiac regulatory score for every base pair that falls within each enhancer. To generate these scores, we have combined the Dickel et al. enhancer predictions with transcription factor ChIP-seq and DNase Hypersensitivity data from human and mouse heart, along with sequence conservation and gene proximity information, as shown below:
This session includes enhancers that: a) differ between normal adult hearts and fetal hearts and b) differ between DCM (dilated cardiomyopathy) adult hearts and normal adult hearts. Enhancer predictions were generated by performing H3K27ac ChIP-seq on adult left ventricular tissue (normal and DCM cohort) and on fetal heart tissue. We compared epigenomic profiles using DiffBind to identify enhancer peaks that are quantitatively different between a) normal adult heart and fetal heart and b) DCM adult heart and normal adult heart. See Spurrell CH, et al. (2019).
This session shows results from the VISTA Enhancer Browser, the largest public collection of sequences tested for enhancer activity in transgenic mouse assays. To date, more than 2,000 human and mouse loci have been tested in vivo at embryonic day 11.5. More than 1,200 show reproducible enhancer activity in at least one tissue, with >200 active in the heart.